AroA
- Description: 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase / chorismate mutase-isozyme 3
| Gene name | aroA |
| Synonyms | aroG |
| Essential | no |
| Product | 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase /
chorismate mutase-isozyme 3 |
| Function | biosynthesis of aromatic amino acids |
| Gene expression levels in SubtiExpress: aroA | |
| Metabolic function and regulation of this protein in SubtiPathways: Phe, Tyr, Trp | |
| MW, pI | 39 kDa, 5.341 |
| Gene length, protein length | 1074 bp, 358 aa |
| Immediate neighbours | ccpA, ytxJ |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 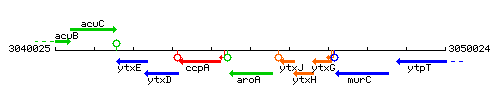
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed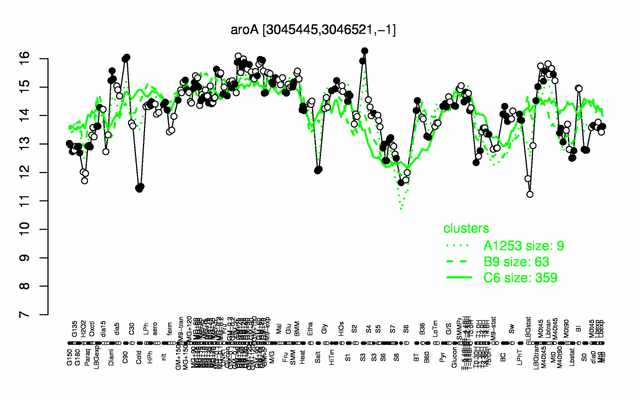
| |
Contents
Categories containing this gene/protein
biosynthesis/ acquisition of amino acids, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU29750
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Phosphoenolpyruvate + D-erythrose 4-phosphate + H2O = 3-deoxy-D-arabino-hept-2-ulosonate 7-phosphate + phosphate (according to Swiss-Prot)
- Protein family: class-I DAHP synthetase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity: subject to feedback inhibition PubMed
Database entries
- Structure: 1VR6 (from Thermotoga maritima, 51% identity, 68% similarity)
- UniProt: P39912
- KEGG entry: [3]
- E.C. number: 2.5.1.54 5 and 5.4.99.5
Additional information
Expression and regulation
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information: subject to feedback inhibition PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Bui Khanh Chi, Alexandra A Roberts, Tran Thi Thanh Huyen, Katrin Bäsell, Dörte Becher, Dirk Albrecht, Chris J Hamilton, Haike Antelmann
S-bacillithiolation protects conserved and essential proteins against hypochlorite stress in firmicutes bacteria.
Antioxid Redox Signal: 2013, 18(11);1273-95
[PubMed:22938038]
[WorldCat.org]
[DOI]
(I p)
Alexander K W Elsholz, Kürsad Turgay, Stephan Michalik, Bernd Hessling, Katrin Gronau, Dan Oertel, Ulrike Mäder, Jörg Bernhardt, Dörte Becher, Michael Hecker, Ulf Gerth
Global impact of protein arginine phosphorylation on the physiology of Bacillus subtilis.
Proc Natl Acad Sci U S A: 2012, 109(19);7451-6
[PubMed:22517742]
[WorldCat.org]
[DOI]
(I p)
Ana Gutiérrez-Preciado, Tina M Henkin, Frank J Grundy, Charles Yanofsky, Enrique Merino
Biochemical features and functional implications of the RNA-based T-box regulatory mechanism.
Microbiol Mol Biol Rev: 2009, 73(1);36-61
[PubMed:19258532]
[WorldCat.org]
[DOI]
(I p)
Christine Eymann, Dörte Becher, Jörg Bernhardt, Katrin Gronau, Anja Klutzny, Michael Hecker
Dynamics of protein phosphorylation on Ser/Thr/Tyr in Bacillus subtilis.
Proteomics: 2007, 7(19);3509-26
[PubMed:17726680]
[WorldCat.org]
[DOI]
(P p)
Boris Macek, Ivan Mijakovic, Jesper V Olsen, Florian Gnad, Chanchal Kumar, Peter R Jensen, Matthias Mann
The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis.
Mol Cell Proteomics: 2007, 6(4);697-707
[PubMed:17218307]
[WorldCat.org]
[DOI]
(P p)
Alia Lapidus, Nathalie Galleron, Alexei Sorokin, S Dusko Ehrlich
Sequencing and functional annotation of the Bacillus subtilis genes in the 200 kb rrnB-dnaB region.
Microbiology (Reading): 1997, 143 ( Pt 11);3431-3441
[PubMed:9387221]
[WorldCat.org]
[DOI]
(P p)