Difference between revisions of "AhrC"
| Line 123: | Line 123: | ||
=References= | =References= | ||
| + | <pubmed>17020585 17576690 18455186 18007040 18007039 11856827 11305941 9383188 7565595 1312212, </pubmed> | ||
# Van Hoy BE et al. (1990) Characterization of the ''spoIVB'' and ''recN'' loci of ''Bacillus subtilis'' ''J Bacteriol.'' '''172:''' 1306-1311. [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=+2106508 PubMed] | # Van Hoy BE et al. (1990) Characterization of the ''spoIVB'' and ''recN'' loci of ''Bacillus subtilis'' ''J Bacteriol.'' '''172:''' 1306-1311. [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=+2106508 PubMed] | ||
# Heidrich et al. (2006) The small untranslated RNA SR1 from the Bacillus subtilis genome is involved in the regulation of arginine catabolism. Mol. Microbiol. 62: 520-536. [http://www.ncbi.nlm.nih.gov/sites/entrez/17020585 PubMed] | # Heidrich et al. (2006) The small untranslated RNA SR1 from the Bacillus subtilis genome is involved in the regulation of arginine catabolism. Mol. Microbiol. 62: 520-536. [http://www.ncbi.nlm.nih.gov/sites/entrez/17020585 PubMed] | ||
Revision as of 20:50, 8 June 2009
- Description: AhrC represses the genes for arginine biosynthesis and activates the genes for arginine catabolism.
| Gene name | ahrC |
| Synonyms | argR |
| Essential | no |
| Product | transcriptional regulator |
| Function | transcriptional regulator of arginine metabolic genes |
| MW, pI | 16,7 kDa, 5.52 |
| Gene length, protein length | 447 bp, 149 amino acids |
| Immediate neighbours | yqxC, recN |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 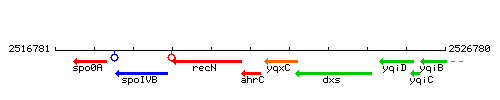
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU24250
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: transcriptional activator/ repressor of genes involved in arginine metabolism
- Protein family: argR family (according to Swiss-Prot)
- Paralogous protein(s):
Genes controlled by AhrC
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s): L-arginine is the co-factor required for transcription repression/ activation
- Effectors of protein activity:
- Interactions:
- Localization: Cytoplasm
Database entries
- Structure: 2P5M (C-Terminus), 2P5K (complex with an 18bp DNA operator), 2P5L (N-terminus), 1F9N, NCBI PubMed,N-Terminus NCBI, C-Terminus NCBI, complex with an 18bp DNA operator NCBI
- Swiss prot entry: P17893
- KEGG entry: [3]
Additional information
Expression and regulation
- Operon: ahrC-recN
- Sigma factor:
- Regulation: by small RNA sr1
- Additional information:
Biological materials
- Mutant: GP729 (aphA3), available in Stülke lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- Antibody:
Labs working on this gene/protein
Simon Phillips, Leeds University, UK Homepage
Michel Debarbouille, Pasteur Institute, Paris, France Homepage
Your additional remarks
References
James A Garnett, Ferenc Marincs, Simon Baumberg, Peter G Stockley, Simon E V Phillips
Structure and function of the arginine repressor-operator complex from Bacillus subtilis.
J Mol Biol: 2008, 379(2);284-98
[PubMed:18455186]
[WorldCat.org]
[DOI]
(I p)
James A Garnett, Simon Baumberg, Peter G Stockley, Simon E V Phillips
Structure of the C-terminal effector-binding domain of AhrC bound to its corepressor L-arginine.
Acta Crystallogr Sect F Struct Biol Cryst Commun: 2007, 63(Pt 11);918-21
[PubMed:18007040]
[WorldCat.org]
[DOI]
(I p)
James A Garnett, Simon Baumberg, Peter G Stockley, Simon E V Phillips
A high-resolution structure of the DNA-binding domain of AhrC, the arginine repressor/activator protein from Bacillus subtilis.
Acta Crystallogr Sect F Struct Biol Cryst Commun: 2007, 63(Pt 11);914-7
[PubMed:18007039]
[WorldCat.org]
[DOI]
(I p)
Nadja Heidrich, Isabella Moll, Sabine Brantl
In vitro analysis of the interaction between the small RNA SR1 and its primary target ahrC mRNA.
Nucleic Acids Res: 2007, 35(13);4331-46
[PubMed:17576690]
[WorldCat.org]
[DOI]
(I p)
Nadja Heidrich, Alberto Chinali, Ulf Gerth, Sabine Brantl
The small untranslated RNA SR1 from the Bacillus subtilis genome is involved in the regulation of arginine catabolism.
Mol Microbiol: 2006, 62(2);520-36
[PubMed:17020585]
[WorldCat.org]
[DOI]
(P p)
Caitríona A Dennis C, Nicholas M Glykos, Mark R Parsons, Simon E V Phillips
The structure of AhrC, the arginine repressor/activator protein from Bacillus subtilis.
Acta Crystallogr D Biol Crystallogr: 2002, 58(Pt 3);421-30
[PubMed:11856827]
[WorldCat.org]
[DOI]
(P p)
K S Makarova, A A Mironov, M S Gelfand
Conservation of the binding site for the arginine repressor in all bacterial lineages.
Genome Biol: 2001, 2(4);RESEARCH0013
[PubMed:11305941]
[WorldCat.org]
[DOI]
(I p)
C M Miller, S Baumberg, P G Stockley
Operator interactions by the Bacillus subtilis arginine repressor/activator, AhrC: novel positioning and DNA-mediated assembly of a transcriptional activator at catabolic sites.
Mol Microbiol: 1997, 26(1);37-48
[PubMed:9383188]
[WorldCat.org]
[DOI]
(P p)
U Klingel, C M Miller, A K North, P G Stockley, S Baumberg
A binding site for activation by the Bacillus subtilis AhrC protein, a repressor/activator of arginine metabolism.
Mol Gen Genet: 1995, 248(3);329-40
[PubMed:7565595]
[WorldCat.org]
[DOI]
(P p)
L G Czaplewski, A K North, M C Smith, S Baumberg, P G Stockley
Purification and initial characterization of AhrC: the regulator of arginine metabolism genes in Bacillus subtilis.
Mol Microbiol: 1992, 6(2);267-75
[PubMed:1312212]
[WorldCat.org]
[DOI]
(P p)
- Van Hoy BE et al. (1990) Characterization of the spoIVB and recN loci of Bacillus subtilis J Bacteriol. 172: 1306-1311. PubMed
- Heidrich et al. (2006) The small untranslated RNA SR1 from the Bacillus subtilis genome is involved in the regulation of arginine catabolism. Mol. Microbiol. 62: 520-536. PubMed
- Heidrich et al. (2007) In vitro analysis of the interaction between the small RNA SR1 and its primary target ahrC mRNA. Nucl. Acids Res. 35: 4331-4346. PubMed
- Garnett et al. (2008) Structure and function of the arginine repressor-operator complex from Bacillus subtilis. J. Mol. Biol. 379: 284-298. PubMed
- Garnett et al. (2007) Structure of the C-terminal effector-binding domain of AhrC bound to its corepressor L-arginine. Acta Cryst. Sect. F. 63: 918-921. PubMed
- Garnett et al. (2007) A high-resolution structure of the DNA-binding domain of AhrC, the arginine repressor/activator protein from Bacillus subtilis. Acta Cryst. Sect. F. 63: 914-917. PubMed
- Dennis et al. (2002) The structure of AhrC, the arginine repressor/activator protein from Bacillus subtilis. Acta Cryst. Sect. D. 58:421-430. PubMed
- Makarova et al. (2001) Conservation of the binding site for the arginine repressor in all bacterial lineages. Genome Biol. 2: RESEARCH0013. PubMed
- Miller et al. (1997) Operator interactions by the Bacillus subtilis arginine repressor/ activator, AhrC: novel positioning and DNA-mediated assembly of a transcriptional activator at catabolic sites. Mol. Microbiol. 26: 37-48. PubMed
- Klingel et al. (1995) A binding site for activation by the Bacillus subtilis AhrC protein, a repressor/ activator of arginine metabolism. Mol. Gen. Genet. 248:329-340. PubMed
- Czaplewski et al. (1992) Purification and initial characterization of AhrC: the regulator of arginine metabolism genes in Bacillus subtilis. Mol. Microbiol. 6:267-275. PubMed
- North et al. (1989) Nucleotide sequence of a Bacillus subtilis arginine regulatory gene and homology of its product to the Escherichia coli arginine repressor. Gene 80:29-38. 2507400 PubMed