AcoA
- Description: acetoin dehydrogenase E1 component (TPP-dependent alpha subunit)
| Gene name | acoA |
| Synonyms | yfjK |
| Essential | no |
| Product | acetoin dehydrogenase E1 component (TPP-dependent alpha subunit) |
| Function | acetoin utilization |
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism | |
| MW, pI | 35 kDa, 4.912 |
| Gene length, protein length | 999 bp, 333 aa |
| Immediate neighbours | yfjL, acoB |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 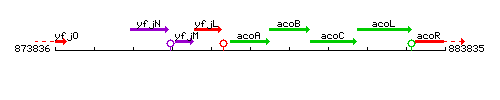
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU08060
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: Cytoplasm (Homogeneous) PubMed
Database entries
- Structure:
- Swiss prot entry: O31404
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism: CcpA: transcription repression, AcoR: transcription activation (interaction with SigL-containing RNA polymerase) PubMed
- Additional information:the mRNA is very stable (half-life > 15 min) PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Michel Debarbouille, Pasteur Institute, Paris, France Homepage
Your additional remarks
References
Jean-Christophe Meile, Ling Juan Wu, S Dusko Ehrlich, Jeff Errington, Philippe Noirot
Systematic localisation of proteins fused to the green fluorescent protein in Bacillus subtilis: identification of new proteins at the DNA replication factory.
Proteomics: 2006, 6(7);2135-46
[PubMed:16479537]
[WorldCat.org]
[DOI]
(P p)
G Hambraeus, C von Wachenfeldt, L Hederstedt
Genome-wide survey of mRNA half-lives in Bacillus subtilis identifies extremely stable mRNAs.
Mol Genet Genomics: 2003, 269(5);706-14
[PubMed:12884008]
[WorldCat.org]
[DOI]
(P p)
N O Ali, J Bignon, G Rapoport, M Debarbouille
Regulation of the acetoin catabolic pathway is controlled by sigma L in Bacillus subtilis.
J Bacteriol: 2001, 183(8);2497-504
[PubMed:11274109]
[WorldCat.org]
[DOI]
(P p)
M Huang, F B Oppermann-Sanio, A Steinbüchel
Biochemical and molecular characterization of the Bacillus subtilis acetoin catabolic pathway.
J Bacteriol: 1999, 181(12);3837-41
[PubMed:10368162]
[WorldCat.org]
[DOI]
(P p)