FapR
Revision as of 13:42, 2 April 2014 by 134.76.38.147 (talk)
- Description: repressor of fatty acid synthetic genes
| Gene name | fapR |
| Synonyms | ylpC |
| Essential | no |
| Product | transcriptional repressor |
| Function | regulation of fatty acid biosynthesis |
| Gene expression levels in SubtiExpress: fapR | |
| Interactions involving this protein in SubtInteract: FapR | |
| Metabolic function and regulation of this protein in SubtiPathways: fapR | |
| MW, pI | 21 kDa, 5.393 |
| Gene length, protein length | 564 bp, 188 aa |
| Immediate neighbours | recG, plsX |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 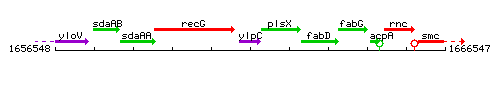
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
biosynthesis of lipids, transcription factors and their control
This gene is a member of the following regulons
The FapR regulon
The gene
Basic information
- Locus tag: BSU15880
Phenotypes of a mutant
Database entries
- BsubCyc: BSU15880
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: FapR regulates the expression of at least 10 genes (the fap regulon). It autoregulates its own expression. Malonyl-CoA, a precursor of fatty acid biosynthesis, binds to FapR changing its conformation to a non-DNA binding state. Hence, conditions that cause malonyl-CoA accumulation, like fatty acid biosynthesis inhibition, derepress the fap regulon.
- Protein family: fapR family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity: malonyl-CoA and malonyl-ACP act as the molecular inducer of the FapR regulon PubMed
Database entries
- BsubCyc: BSU15880
- UniProt: O34835
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
The FapR regulon
Gustavo E Schujman, Luciana Paoletti, Alan D Grossman, Diego de Mendoza
FapR, a bacterial transcription factor involved in global regulation of membrane lipid biosynthesis.
Dev Cell: 2003, 4(5);663-72
[PubMed:12737802]
[WorldCat.org]
[DOI]
(P p)
Other original publications
Additional publications: PubMed