XhlA
- Description: involved in cell lysis upon induction of PBSX
| Gene name | xhlA |
| Synonyms | |
| Essential | no |
| Product | unknown |
| Function | host cell lysis |
| MW, pI | 9 kDa, 5.073 |
| Gene length, protein length | 267 bp, 89 aa |
| Immediate neighbours | xepA, xhlB |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 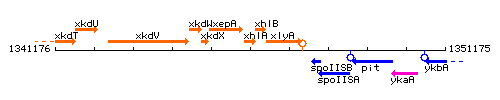
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
PBSX prophage, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU12790
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- XhlA is a member of a suspected group of hubs proteins that were suggested to be involved in a large number of interactions PubMed
- Localization: cell membrane (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: P39798
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Elodie Marchadier, Rut Carballido-López, Sophie Brinster, Céline Fabret, Peggy Mervelet, Philippe Bessières, Marie-Françoise Noirot-Gros, Vincent Fromion, Philippe Noirot
An expanded protein-protein interaction network in Bacillus subtilis reveals a group of hubs: Exploration by an integrative approach.
Proteomics: 2011, 11(15);2981-91
[PubMed:21630458]
[WorldCat.org]
[DOI]
(I p)
S Krogh, S T Jørgensen, K M Devine
Lysis genes of the Bacillus subtilis defective prophage PBSX.
J Bacteriol: 1998, 180(8);2110-7
[PubMed:9555893]
[WorldCat.org]
[DOI]
(P p)
P F Longchamp, C Mauël, D Karamata
Lytic enzymes associated with defective prophages of Bacillus subtilis: sequencing and characterization of the region comprising the N-acetylmuramoyl-L-alanine amidase gene of prophage PBSX.
Microbiology (Reading): 1994, 140 ( Pt 8);1855-67
[PubMed:7921239]
[WorldCat.org]
[DOI]
(P p)