YyaF
- Description: GTP-binding protein/ GTPase
| Gene name | yyaF |
| Synonyms | |
| Essential | no |
| Product | GTP-binding protein/ GTPase |
| Function | unknown |
| Gene expression levels in SubtiExpress: yyaF | |
| MW, pI | 39 kDa, 4.659 |
| Gene length, protein length | 1098 bp, 366 aa |
| Immediate neighbours | rpsF, yyaE |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 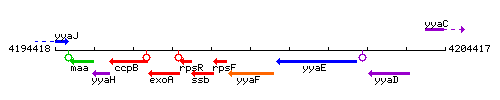
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed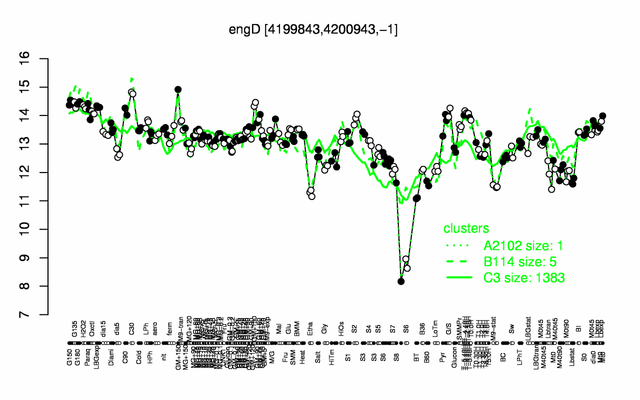
| |
Contents
Categories containing this gene/protein
GTP-binding proteins, proteins of unknown function
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU40920
Phenotypes of a mutant
Database entries
- BsubCyc: BSU40920
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Binds and hydrolyzes GTP and readily exchanges GDP for GTP
- Protein family: EngD subfamily (according to Swiss-Prot) Era/Obg family
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- BsubCyc: BSU40920
- Structure:
- UniProt: P37518
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Sigma factor:
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Naotake Ogasawara, Nara, Japan
Your additional remarks
References
Reviews
Natalie Verstraeten, Maarten Fauvart, Wim Versées, Jan Michiels
The universally conserved prokaryotic GTPases.
Microbiol Mol Biol Rev: 2011, 75(3);507-42, second and third pages of table of contents
[PubMed:21885683]
[WorldCat.org]
[DOI]
(I p)
Original publications
Cordula Lindner, Reindert Nijland, Mariska van Hartskamp, Sierd Bron, Leendert W Hamoen, Oscar P Kuipers
Differential expression of two paralogous genes of Bacillus subtilis encoding single-stranded DNA binding protein.
J Bacteriol: 2004, 186(4);1097-105
[PubMed:14762004]
[WorldCat.org]
[DOI]
(P p)
Takuya Morimoto, Pek Chin Loh, Tomohiro Hirai, Kei Asai, Kazuo Kobayashi, Shigeki Moriya, Naotake Ogasawara
Six GTP-binding proteins of the Era/Obg family are essential for cell growth in Bacillus subtilis.
Microbiology (Reading): 2002, 148(Pt 11);3539-3552
[PubMed:12427945]
[WorldCat.org]
[DOI]
(P p)
Christine Eymann, Georg Homuth, Christian Scharf, Michael Hecker
Bacillus subtilis functional genomics: global characterization of the stringent response by proteome and transcriptome analysis.
J Bacteriol: 2002, 184(9);2500-20
[PubMed:11948165]
[WorldCat.org]
[DOI]
(P p)
Mitsuo Ogura, Hirotake Yamaguchi, Kazuo Kobayashi, Naotake Ogasawara, Yasutaro Fujita, Teruo Tanaka
Whole-genome analysis of genes regulated by the Bacillus subtilis competence transcription factor ComK.
J Bacteriol: 2002, 184(9);2344-51
[PubMed:11948146]
[WorldCat.org]
[DOI]
(P p)