Difference between revisions of "DhbF"
| Line 120: | Line 120: | ||
=References= | =References= | ||
| − | <pubmed>18763711 17218307, </pubmed> | + | <pubmed>12354229,18763711 17218307, </pubmed> |
# Hahne et al. (2008) From complementarity to comprehensiveness - targeting the membrane proteome of growing ''Bacillus subtilis'' by divergent approaches. Proteomics '''8:''' 4123-4136 [http://www.ncbi.nlm.nih.gov/pubmed/18763711 PubMed] | # Hahne et al. (2008) From complementarity to comprehensiveness - targeting the membrane proteome of growing ''Bacillus subtilis'' by divergent approaches. Proteomics '''8:''' 4123-4136 [http://www.ncbi.nlm.nih.gov/pubmed/18763711 PubMed] | ||
# Macek et al. (2007) The serine/ threonine/ tyrosine phosphoproteome of the model bacterium ''Bacillus subtilis''. Mol. Cell. Proteomics 6: 697-707 [http://www.ncbi.nlm.nih.gov/pubmed/17218307 PubMed] | # Macek et al. (2007) The serine/ threonine/ tyrosine phosphoproteome of the model bacterium ''Bacillus subtilis''. Mol. Cell. Proteomics 6: 697-707 [http://www.ncbi.nlm.nih.gov/pubmed/17218307 PubMed] | ||
# Author1, Author2 & Author3 (year) Title ''Journal'' '''volume:''' page-page. [http://www.ncbi.nlm.nih.gov/sites/entrez/PMID PubMed] | # Author1, Author2 & Author3 (year) Title ''Journal'' '''volume:''' page-page. [http://www.ncbi.nlm.nih.gov/sites/entrez/PMID PubMed] | ||
Revision as of 12:11, 12 June 2009
- Description: involved in 2,3-dihydroxybenzoate biosynthesis
| Gene name | dhbF |
| Synonyms | |
| Essential | no |
| Product | unknown |
| Function | siderophore biosynthesis |
| MW, pI | 263 kDa, 4.737 |
| Gene length, protein length | 7134 bp, 2378 aa |
| Immediate neighbours | yukJ, dhbB |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 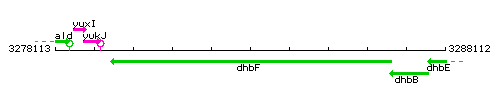
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU31960
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: ATP-dependent AMP-binding enzyme family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification: phosphorylation on Ser-988 AND Ser-996 PubMed
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: membrane PubMed
Database entries
- Structure:
- Swiss prot entry: P45745
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
- Hahne et al. (2008) From complementarity to comprehensiveness - targeting the membrane proteome of growing Bacillus subtilis by divergent approaches. Proteomics 8: 4123-4136 PubMed
- Macek et al. (2007) The serine/ threonine/ tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol. Cell. Proteomics 6: 697-707 PubMed
- Author1, Author2 & Author3 (year) Title Journal volume: page-page. PubMed