Difference between revisions of "YtsJ"
| Line 80: | Line 80: | ||
* '''Structure:''' | * '''Structure:''' | ||
| − | * '''Swiss prot entry:''' | + | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/O34962 O34962] |
* '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+BSU29220] | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+BSU29220] | ||
Revision as of 09:12, 5 May 2009
- Description: malic enzyme
| Gene name | ytsJ |
| Synonyms | |
| Essential | no |
| Product | NAD-dependent malate dehydrogenase |
| Function | malate utilization |
| MW, pI | 43 kDa, 5.046 |
| Gene length, protein length | 1230 bp, 410 aa |
| Immediate neighbours | accD, dnaE |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 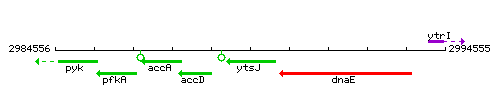
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Coordinates:
Phenotypes of a mutant
Poor growth with malate as single carbon source PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: malate <--> pyruvate
- Protein family:
- Paralogous protein(s): MleA
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
Database entries
- Structure:
- Swiss prot entry: O34962
- KEGG entry: [3]
- E.C. number: 1.1.1.40
Additional information
Expression and regulation
- Operon:
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant: GP612 (spc), available in Stülke lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Stephane Aymerich, Microbiology and Molecular Genetics, INRA Paris-Grignon, France
Your additional remarks
References
- Meile et al. (2006) Systematic localisation of proteins fused to the green fluorescent protein in Bacillus subtilis: identification of new proteins at the DNA replication factory Proteomics 6: 2135-2146. PubMed# Meile et al. (2006) Systematic localisation of proteins fused to the green fluorescent protein in Bacillus subtilis: identification of new proteins at the DNA replication factory Proteomics 6: 2135-2146. PubMed
- Lerondel G, Doan T, Zamboni N, Sauer U, Aymerich S. (2006) YtsJ has the major physiological role of the four paralogous malic enzyme isoforms in Bacillus subtilis. J Bacteriol. 188: 4727-4736. PubMed
- Doan, T., P. Servant, S. Tojo, H. Yamaguchi, G. Lerondel, K. Yoshida, Y. Fujita, and S. Aymerich. 2003. The Bacillus subtilis ywkA gene encodes a malic enzyme and its transcription is activated by the YufL/YufM two-component system in response to malate. Microbiology 149: 2331-2343. PubMed
- Author1, Author2 & Author3 (year) Title Journal volume: page-page. PubMed