Difference between revisions of "DnaK"
| Line 128: | Line 128: | ||
** number of protein molecules per cell (minimal medium with glucose and ammonium): 5860 {{PubMed|24696501}} | ** number of protein molecules per cell (minimal medium with glucose and ammonium): 5860 {{PubMed|24696501}} | ||
** number of protein molecules per cell (complex medium with amino acids, without glucose): 22949 {{PubMed|24696501}} | ** number of protein molecules per cell (complex medium with amino acids, without glucose): 22949 {{PubMed|24696501}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 8450 {{PubMed|21395229}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 4859 {{PubMed|21395229}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 2421 {{PubMed|21395229}} | ||
=Biological materials = | =Biological materials = | ||
| − | |||
* '''Mutant:''' | * '''Mutant:''' | ||
Revision as of 14:05, 17 April 2014
- Description: class I heat-shock protein (molecular chaperone)
| Gene name | dnaK |
| Synonyms | |
| Essential | no |
| Product | class I heat-shock protein (molecular chaperone) |
| Function | protein quality control |
| Gene expression levels in SubtiExpress: dnaK | |
| Interactions involving this protein in SubtInteract: DnaK | |
| Metabolic function and regulation of this protein in SubtiPathways: dnaK | |
| MW, pI | 65 kDa, 4.571 |
| Gene length, protein length | 1833 bp, 611 aa |
| Immediate neighbours | surC, grpE |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 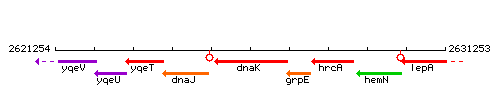
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed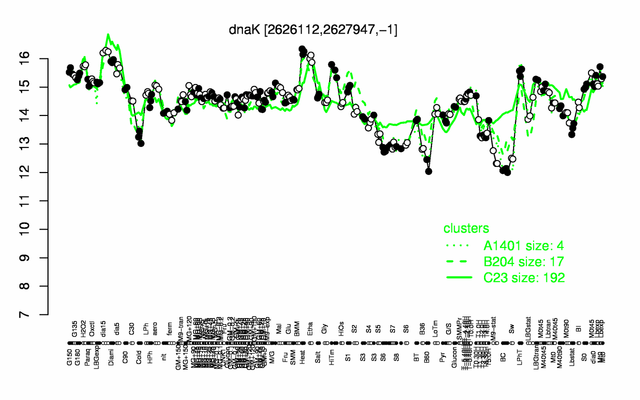
| |
Contents
Categories containing this gene/protein
chaperones/ protein folding, heat shock proteins, phosphoproteins, most abundant proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU25470
Phenotypes of a mutant
Database entries
- BsubCyc: BSU25470
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: heat shock protein 70 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification: phosphorylated on ser/ thr/ tyr PubMed
- Effectors of protein activity:
Database entries
- BsubCyc: BSU25470
- Structure:
- UniProt: P17820
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
- belongs to the 100 most abundant proteins PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium): 5860 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 22949 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 8450 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 4859 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 2421 PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody:
Labs working on this gene/protein
Wolfgang Schumann, Bayreuth University, Germany Homepage
Your additional remarks
References
G Seydlová, P Halada, R Fišer, O Toman, A Ulrych, J Svobodová
DnaK and GroEL chaperones are recruited to the Bacillus subtilis membrane after short-term ethanol stress.
J Appl Microbiol: 2012, 112(4);765-74
[PubMed:22268681]
[WorldCat.org]
[DOI]
(I p)
Christine Eymann, Dörte Becher, Jörg Bernhardt, Katrin Gronau, Anja Klutzny, Michael Hecker
Dynamics of protein phosphorylation on Ser/Thr/Tyr in Bacillus subtilis.
Proteomics: 2007, 7(19);3509-26
[PubMed:17726680]
[WorldCat.org]
[DOI]
(P p)
Jean-Christophe Meile, Ling Juan Wu, S Dusko Ehrlich, Jeff Errington, Philippe Noirot
Systematic localisation of proteins fused to the green fluorescent protein in Bacillus subtilis: identification of new proteins at the DNA replication factory.
Proteomics: 2006, 6(7);2135-46
[PubMed:16479537]
[WorldCat.org]
[DOI]
(P p)
Christine Eymann, Annette Dreisbach, Dirk Albrecht, Jörg Bernhardt, Dörte Becher, Sandy Gentner, Le Thi Tam, Knut Büttner, Gerrit Buurman, Christian Scharf, Simone Venz, Uwe Völker, Michael Hecker
A comprehensive proteome map of growing Bacillus subtilis cells.
Proteomics: 2004, 4(10);2849-76
[PubMed:15378759]
[WorldCat.org]
[DOI]
(P p)
Dindo Y Reyes, Hirofumi Yoshikawa
DnaK chaperone machine and trigger factor are only partially required for normal growth of Bacillus subtilis.
Biosci Biotechnol Biochem: 2002, 66(7);1583-6
[PubMed:12224648]
[WorldCat.org]
[DOI]
(P p)
G Homuth, A Mogk, W Schumann
Post-transcriptional regulation of the Bacillus subtilis dnaK operon.
Mol Microbiol: 1999, 32(6);1183-97
[PubMed:10383760]
[WorldCat.org]
[DOI]
(P p)
G Homuth, S Masuda, A Mogk, Y Kobayashi, W Schumann
The dnaK operon of Bacillus subtilis is heptacistronic.
J Bacteriol: 1997, 179(4);1153-64
[PubMed:9023197]
[WorldCat.org]
[DOI]
(P p)
G Yuan, S L Wong
Isolation and characterization of Bacillus subtilis groE regulatory mutants: evidence for orf39 in the dnaK operon as a repressor gene in regulating the expression of both groE and dnaK.
J Bacteriol: 1995, 177(22);6462-8
[PubMed:7592421]
[WorldCat.org]
[DOI]
(P p)
A Schulz, B Tzschaschel, W Schumann
Isolation and analysis of mutants of the dnaK operon of Bacillus subtilis.
Mol Microbiol: 1995, 15(3);421-9
[PubMed:7540247]
[WorldCat.org]
[DOI]
(P p)
M Wetzstein, U Völker, J Dedio, S Löbau, U Zuber, M Schiesswohl, C Herget, M Hecker, W Schumann
Cloning, sequencing, and molecular analysis of the dnaK locus from Bacillus subtilis.
J Bacteriol: 1992, 174(10);3300-10
[PubMed:1339421]
[WorldCat.org]
[DOI]
(P p)