Difference between revisions of "RpoY"
| Line 114: | Line 114: | ||
* '''Additional information:''' | * '''Additional information:''' | ||
** translation of [[YkzG]] and [[RnjA]] is coupled, and this coupling is required for efficient expression of [[rnjA|RNase J1]] {{PubMed|24187087}} | ** translation of [[YkzG]] and [[RnjA]] is coupled, and this coupling is required for efficient expression of [[rnjA|RNase J1]] {{PubMed|24187087}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium): 443 {{PubMed|24696501}} | ||
=Biological materials = | =Biological materials = | ||
Revision as of 10:04, 17 April 2014
- Description: epsilon subunit of RNA polymerase
| Gene name | ykzG |
| Synonyms | |
| Essential | no |
| Product | epsilon subunit of RNA polymerase |
| Function | control of RNA polymerase activity |
| Gene expression levels in SubtiExpress: ykzG | |
| MW, pI | 8 kDa, 4.605 |
| Gene length, protein length | 207 bp, 69 aa |
| Immediate neighbours | rnjA, ykrA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 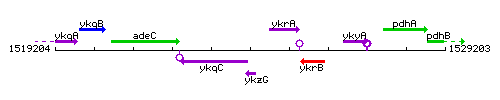
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU14540
Phenotypes of a mutant
Database entries
- BsubCyc: BSU14540
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein is sometimes annotated as omega1 subunit of RNA polymerase, however, there is no experimental evidence in support of this ("it is unrelated in sequence to ω/Rpb6 from the eubacteria, archaea and eukaryotes, unpublished data suggests it does not bind at the same site as ω/Rpb6 subunits bind their cognate RNAPs, and therefore it is unlikely to function as a true ω subunit") PubMed
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: UPF0356 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- BsubCyc: BSU14540
- Structure:
- UniProt: O31718
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Ailar Jamalli, Agnès Hébert, Léna Zig, Harald Putzer
Control of expression of the RNases J1 and J2 in Bacillus subtilis.
J Bacteriol: 2014, 196(2);318-24
[PubMed:24187087]
[WorldCat.org]
[DOI]
(I p)
Geoff P Doherty, Mark J Fogg, Anthony J Wilkinson, Peter J Lewis
Small subunits of RNA polymerase: localization, levels and implications for core enzyme composition.
Microbiology (Reading): 2010, 156(Pt 12);3532-3543
[PubMed:20724389]
[WorldCat.org]
[DOI]
(I p)