Difference between revisions of "BrxA"
| Line 35: | Line 35: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | |||
| − | |||
| − | |||
| − | |||
<br/><br/> | <br/><br/> | ||
| Line 88: | Line 84: | ||
* '''[[SubtInteract|Interactions]]:''' | * '''[[SubtInteract|Interactions]]:''' | ||
| − | * '''Localization:''' | + | * '''[[Localization]]:''' |
=== Database entries === | === Database entries === | ||
| Line 108: | Line 104: | ||
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=yphP_2300221_2300655_-1 yphP] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=yphP_2300221_2300655_-1 yphP] {{PubMed|22383849}} | ||
| − | * '''Sigma factor:''' | + | * '''[[Sigma factor]]:''' |
* '''Regulation:''' | * '''Regulation:''' | ||
Revision as of 19:12, 18 June 2013
- Description: disulfide isomerase, putative bacilliredoxin
| Gene name | yphP |
| Synonyms | |
| Essential | no |
| Product | disulfide isomerase, putative bacilliredoxin |
| Function | unknown |
| Gene expression levels in SubtiExpress: yphP | |
| MW, pI | 15 kDa, 4.619 |
| Gene length, protein length | 432 bp, 144 aa |
| Immediate neighbours | ypiP, ilvD |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 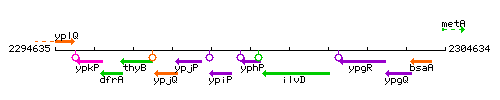
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed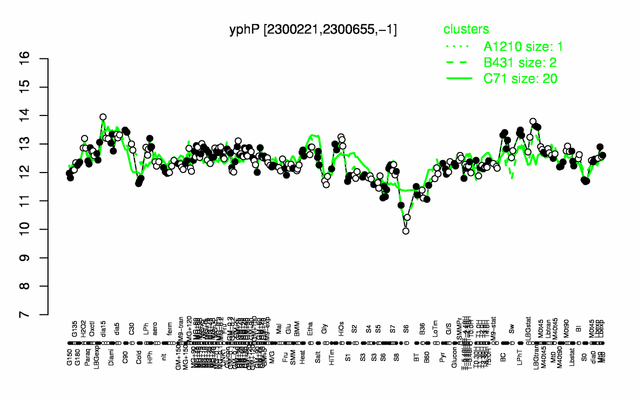
| |
Contents
Categories containing this gene/protein
poorly characterized/ putative enzymes
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU21860
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: scpA family (according to Swiss-Prot)
- Paralogous protein(s): yqiW
Extended information on the protein
- Kinetic information:
- Domains: contains an CxC redox motif [3]
- Modification:
- S-bacillithiolation upon hypochlorite stress on Cys-53 PubMed
- Cofactor(s):
- Effectors of protein activity:
Database entries
- UniProt: P54170
- KEGG entry: [4]
- E.C. number:
Additional information
Expression and regulation
- Operon: yphP (according to DBTBS)
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
YpdA, YqiW, YphP and YtxJ co-occur in those species predicted to synthesize bacillithiol.
References
Bui Khanh Chi, Alexandra A Roberts, Tran Thi Thanh Huyen, Katrin Bäsell, Dörte Becher, Dirk Albrecht, Chris J Hamilton, Haike Antelmann
S-bacillithiolation protects conserved and essential proteins against hypochlorite stress in firmicutes bacteria.
Antioxid Redox Signal: 2013, 18(11);1273-95
[PubMed:22938038]
[WorldCat.org]
[DOI]
(I p)
Bui Khanh Chi, Katrin Gronau, Ulrike Mäder, Bernd Hessling, Dörte Becher, Haike Antelmann
S-bacillithiolation protects against hypochlorite stress in Bacillus subtilis as revealed by transcriptomics and redox proteomics.
Mol Cell Proteomics: 2011, 10(11);M111.009506
[PubMed:21749987]
[WorldCat.org]
[DOI]
(I p)
Ahmed Gaballa, Gerald L Newton, Haike Antelmann, Derek Parsonage, Heather Upton, Mamta Rawat, Al Claiborne, Robert C Fahey, John D Helmann
Biosynthesis and functions of bacillithiol, a major low-molecular-weight thiol in Bacilli.
Proc Natl Acad Sci U S A: 2010, 107(14);6482-6
[PubMed:20308541]
[WorldCat.org]
[DOI]
(I p)
Urszula Derewenda, Tomasz Boczek, Kelly L Gorres, Minmin Yu, Li-wei Hung, David Cooper, Andrzej Joachimiak, Ronald T Raines, Zygmunt S Derewenda
Structure and function of Bacillus subtilis YphP, a prokaryotic disulfide isomerase with a CXC catalytic motif .
Biochemistry: 2009, 48(36);8664-71
[PubMed:19653655]
[WorldCat.org]
[DOI]
(I p)