Difference between revisions of "FolE"
| Line 29: | Line 29: | ||
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
|- | |- | ||
| − | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=folEA_2384783_2385355_-1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:folE_expression.png|500px]] | + | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=folEA_2384783_2385355_-1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:folE_expression.png|500px|link=http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU22780]] |
|- | |- | ||
|} | |} | ||
Revision as of 13:45, 16 May 2013
- Description: GTP cyclohydrolase I
| Gene name | folE |
| Synonyms | mtrA |
| Essential | no |
| Product | GTP cyclohydrolase IA |
| Function | biosynthesis of folate |
| Gene expression levels in SubtiExpress: folE | |
| Metabolic function and regulation of this protein in SubtiPathways: Phe, Tyr, Trp, Folate | |
| MW, pI | 21 kDa, 6.335 |
| Gene length, protein length | 570 bp, 190 aa |
| Immediate neighbours | mtrB, hbs |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 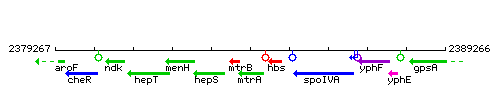
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed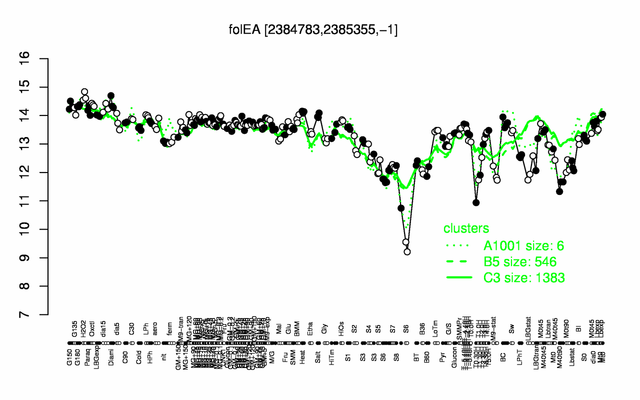
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU22780
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: GTP + H2O = formate + 2-amino-4-hydroxy-6-(erythro-1,2,3-trihydroxypropyl)-dihydropteridine triphosphate (according to Swiss-Prot)
- Protein family: citrate synthase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s): zinc-dependent enzyme PubMed
- Effectors of protein activity:
Database entries
- UniProt: P19465
- KEGG entry: [3]
- E.C. number: 3.5.4.16
Additional information
Expression and regulation
- Regulation: constitutive PubMed
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Banumathi Sankaran, Shilah A Bonnett, Kinjal Shah, Scott Gabriel, Robert Reddy, Paul Schimmel, Dmitry A Rodionov, Valérie de Crécy-Lagard, John D Helmann, Dirk Iwata-Reuyl, Manal A Swairjo
Zinc-independent folate biosynthesis: genetic, biochemical, and structural investigations reveal new metal dependence for GTP cyclohydrolase IB.
J Bacteriol: 2009, 191(22);6936-49
[PubMed:19767425]
[WorldCat.org]
[DOI]
(I p)
Basma El Yacoubi, Shilah Bonnett, Jessica N Anderson, Manal A Swairjo, Dirk Iwata-Reuyl, Valérie de Crécy-Lagard
Discovery of a new prokaryotic type I GTP cyclohydrolase family.
J Biol Chem: 2006, 281(49);37586-93
[PubMed:17032654]
[WorldCat.org]
[DOI]
(P p)
P Babitzke, P Gollnick, C Yanofsky
The mtrAB operon of Bacillus subtilis encodes GTP cyclohydrolase I (MtrA), an enzyme involved in folic acid biosynthesis, and MtrB, a regulator of tryptophan biosynthesis.
J Bacteriol: 1992, 174(7);2059-64
[PubMed:1551827]
[WorldCat.org]
[DOI]
(P p)
P Gollnick, S Ishino, M I Kuroda, D J Henner, C Yanofsky
The mtr locus is a two-gene operon required for transcription attenuation in the trp operon of Bacillus subtilis.
Proc Natl Acad Sci U S A: 1990, 87(22);8726-30
[PubMed:2123343]
[WorldCat.org]
[DOI]
(P p)