Difference between revisions of "PurF"
| Line 23: | Line 23: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[purL]]'', ''[[purM]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[purL]]'', ''[[purM]]'' | ||
|- | |- | ||
| − | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU06490 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU06490 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU06490 | + | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU06490 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU06490 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU06490 DNA_with_flanks] |
|- | |- | ||
|- | |- | ||
Revision as of 09:37, 14 May 2013
- Description: glutamine phosphoribosyldiphosphate amidotransferase
| Gene name | purF |
| Synonyms | purB |
| Essential | no |
| Product | glutamine phosphoribosyldiphosphate amidotransferase |
| Function | purine biosynthesis |
| Gene expression levels in SubtiExpress: purF | |
| Metabolic function and regulation of this protein in SubtiPathways: Purine synthesis, Nucleotides (regulation) | |
| MW, pI | 51 kDa, 5.873 |
| Gene length, protein length | 1428 bp, 476 aa |
| Immediate neighbours | purL, purM |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 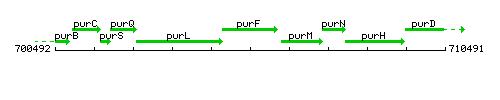
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed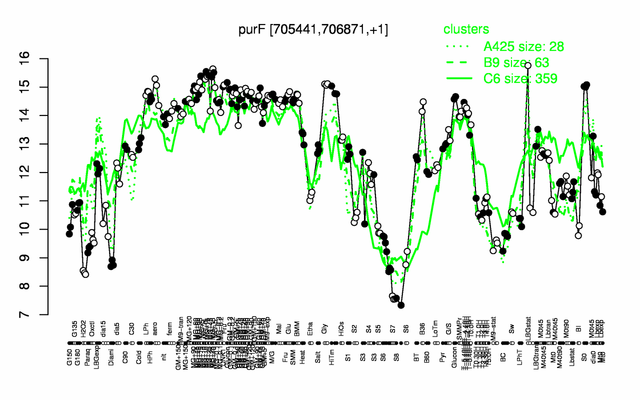
| |
Contents
Categories containing this gene/protein
biosynthesis/ acquisition of nucleotides
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU06490
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 5-phospho-beta-D-ribosylamine + diphosphate + L-glutamate = L-glutamine + 5-phospho-alpha-D-ribose 1-diphosphate + H2O (according to Swiss-Prot)
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- UniProt: P00497
- KEGG entry: [3]
- E.C. number: 2.4.2.14
Additional information
- subject to Clp-dependent proteolysis upon glucose starvation PubMed
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information: subject to Clp-dependent proteolysis upon glucose starvation PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Lars Engholm Johansen, Per Nygaard, Catharina Lassen, Yvonne Agersø, Hans H Saxild
Definition of a second Bacillus subtilis pur regulon comprising the pur and xpt-pbuX operons plus pbuG, nupG (yxjA), and pbuE (ydhL).
J Bacteriol: 2003, 185(17);5200-9
[PubMed:12923093]
[WorldCat.org]
[DOI]
(P p)
M Weng, P L Nagy, H Zalkin
Identification of the Bacillus subtilis pur operon repressor.
Proc Natl Acad Sci U S A: 1995, 92(16);7455-9
[PubMed:7638212]
[WorldCat.org]
[DOI]
(P p)
J L Smith, E J Zaluzec, J P Wery, L Niu, R L Switzer, H Zalkin, Y Satow
Structure of the allosteric regulatory enzyme of purine biosynthesis.
Science: 1994, 264(5164);1427-33
[PubMed:8197456]
[WorldCat.org]
[DOI]
(P p)
Y A Oñate, S J Vollmer, R L Switzer, M K Johnson
Spectroscopic characterization of the iron-sulfur cluster in Bacillus subtilis glutamine phosphoribosylpyrophosphate amidotransferase.
J Biol Chem: 1989, 264(31);18386-91
[PubMed:2553706]
[WorldCat.org]
(P p)
J A Grandoni, R L Switzer, C A Makaroff, H Zalkin
Evidence that the iron-sulfur cluster of Bacillus subtilis glutamine phosphoribosylpyrophosphate amidotransferase determines stability of the enzyme to degradation in vivo.
J Biol Chem: 1989, 264(11);6058-64
[PubMed:2495277]
[WorldCat.org]
(P p)
D J Ebbole, H Zalkin
Cloning and characterization of a 12-gene cluster from Bacillus subtilis encoding nine enzymes for de novo purine nucleotide synthesis.
J Biol Chem: 1987, 262(17);8274-87
[PubMed:3036807]
[WorldCat.org]
(P p)
S J Vollmer, R L Switzer, P G Debrunner
Oxidation-reduction properties of the iron-sulfur cluster in Bacillus subtilis glutamine phosphoribosylpyrophosphate amidotransferase.
J Biol Chem: 1983, 258(23);14284-93
[PubMed:6315725]
[WorldCat.org]
(P p)
C A Makaroff, H Zalkin, R L Switzer, S J Vollmer
Cloning of the Bacillus subtilis glutamine phosphoribosylpyrophosphate amidotransferase gene in Escherichia coli. Nucleotide sequence determination and properties of the plasmid-encoded enzyme.
J Biol Chem: 1983, 258(17);10586-93
[PubMed:6411717]
[WorldCat.org]
(P p)
S J Vollmer, R L Switzer, M A Hermodson, S G Bower, H Zalkin
The glutamine-utilizing site of Bacillus subtilis glutamine phosphoribosylpyrophosphate amidotransferase.
J Biol Chem: 1983, 258(17);10582-5
[PubMed:6411716]
[WorldCat.org]
(P p)
D A Bernlohr, R L Switzer
Regulation of Bacillus subtilis glutamine phosphoribosylpyrophosphate amidotransferase inactivation in vivo.
J Bacteriol: 1983, 153(2);937-49
[PubMed:6401710]
[WorldCat.org]
[DOI]
(P p)
R L Switzer, M E Ruppen, D A Bernlohr
Inactivation of glutamine: 5-phosphoribosyl 1-pyrophosphate amidotransferase in Bacillus subtilis: oxidation of an essential Fe-S centre precedes selective degradation.
Biochem Soc Trans: 1982, 10(5);322-4
[PubMed:6814966]
[WorldCat.org]
[DOI]
(P p)
D A Bernlohr, R L Switzer
Reaction of Bacillus subtilis glutamine phosphoribosylpyrophosphate amidotransferase with oxygen: chemistry and regulation by ligands.
Biochemistry: 1981, 20(20);5675-81
[PubMed:6794614]
[WorldCat.org]
[DOI]
(P p)
J Y Wong, D A Bernlohr, C L Turnbough, R L Switzer
Purification and properties of glutamine phosphoribosylpyrophosphate amidotransferase from Bacillus subtilis.
Biochemistry: 1981, 20(20);5669-74
[PubMed:6794613]
[WorldCat.org]
[DOI]
(P p)
B A Averill, A Dwivedi, P Debrunner, S J Vollmer, J Y Wong, R L Switzer
Evidence for a tetranuclear iron-sulfur center in glutamine phosphoribosylpyrophosphate amidotransferase from Bacillus subtilis.
J Biol Chem: 1980, 255(13);6007-10
[PubMed:6771260]
[WorldCat.org]
(P p)
E Meyer, R L Switzer
Regulation of Bacillus subtilis glutamine phosphoribosylpyrophosphate amidotransferase activity by end products.
J Biol Chem: 1979, 254(12);5397-402
[PubMed:109433]
[WorldCat.org]
(P p)