Difference between revisions of "RecG"
m (Reverted edits by 134.76.70.252 (talk) to last revision by Jstuelk) |
|||
| Line 100: | Line 100: | ||
* '''Operon:''' | * '''Operon:''' | ||
| − | * '''[ | + | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=recG_1659810_1661858_1 recG] {{PubMed|22383849}} |
| + | |||
| + | * '''Sigma factor:''' | ||
* '''Regulation:''' | * '''Regulation:''' | ||
| Line 106: | Line 108: | ||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| − | * '''Additional information:''' | + | * '''Additional information:''' |
=Biological materials = | =Biological materials = | ||
Revision as of 08:48, 13 April 2012
- Description: ATP-dependent DNA helicase, branch migration translocase, required for DNA repair and chromosomal segregation
| Gene name | recG |
| Synonyms | ylpB |
| Essential | no |
| Product | ATP-dependent DNA helicase, branch migration translocase |
| Function | DNA repair and chromosomal segregation |
| Interactions involving this protein in SubtInteract: RecG | |
| Metabolic function and regulation of this protein in SubtiPathways: Ala, Gly, Ser | |
| MW, pI | 77 kDa, 7.093 |
| Gene length, protein length | 2046 bp, 682 aa |
| Immediate neighbours | sdaAA, fapR |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 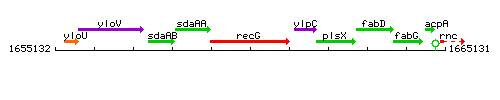
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU15870
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: helicase C-terminal domain (according to Swiss-Prot)
- Paralogous protein(s): Mfd
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure: 1GM5 (from Thermotoga maritima, 43% identity, 62% similarity)
- UniProt: O34942
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
François Lecointe, Céline Sérèna, Marion Velten, Audrey Costes, Stephen McGovern, Jean-Christophe Meile, Jeffrey Errington, S Dusko Ehrlich, Philippe Noirot, Patrice Polard
Anticipating chromosomal replication fork arrest: SSB targets repair DNA helicases to active forks.
EMBO J: 2007, 26(19);4239-51
[PubMed:17853894]
[WorldCat.org]
[DOI]
(P p)
Humberto Sanchez, Begoña Carrasco, M Castillo Cozar, Juan C Alonso
Bacillus subtilis RecG branch migration translocase is required for DNA repair and chromosomal segregation.
Mol Microbiol: 2007, 65(4);920-35
[PubMed:17640277]
[WorldCat.org]
[DOI]
(P p)
Qin Wen, Akeel A Mahdi, Geoffrey S Briggs, Gary J Sharples, Robert G Lloyd
Conservation of RecG activity from pathogens to hyperthermophiles.
DNA Repair (Amst): 2005, 4(1);23-31
[PubMed:15533834]
[WorldCat.org]
[DOI]
(P p)
Additional publications: PubMed