Difference between revisions of "YodC"
| Line 119: | Line 119: | ||
<pubmed>17407181,18208493,, </pubmed> | <pubmed>17407181,18208493,, </pubmed> | ||
| + | |||
| + | [[Category:Protein-coding genes]] | ||
Revision as of 11:10, 21 July 2009
- Description: similar to nitroreductase
| Gene name | yodC |
| Synonyms | |
| Essential | no |
| Product | unknown |
| Function | unknown |
| MW, pI | 22 kDa, 5.274 |
| Gene length, protein length | 606 bp, 202 aa |
| Immediate neighbours | yodB, mhqD |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 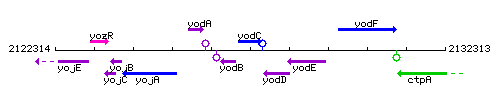
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU19550
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: type A lantibiotic family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure:
- UniProt: P81102
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Montira Leelakriangsak, Nguyen Thi Thu Huyen, Stefanie Töwe, Nguyen van Duy, Dörte Becher, Michael Hecker, Haike Antelmann, Peter Zuber
Regulation of quinone detoxification by the thiol stress sensing DUF24/MarR-like repressor, YodB in Bacillus subtilis.
Mol Microbiol: 2008, 67(5);1108-24
[PubMed:18208493]
[WorldCat.org]
[DOI]
(I p)
Van Duy Nguyen, Carmen Wolf, Ulrike Mäder, Michael Lalk, Peter Langer, Ulrike Lindequist, Michael Hecker, Haike Antelmann
Transcriptome and proteome analyses in response to 2-methylhydroquinone and 6-brom-2-vinyl-chroman-4-on reveal different degradation systems involved in the catabolism of aromatic compounds in Bacillus subtilis.
Proteomics: 2007, 7(9);1391-408
[PubMed:17407181]
[WorldCat.org]
[DOI]
(P p)