Difference between revisions of "Eno"
(→Biological materials) |
(→Biological materials) |
||
| Line 116: | Line 116: | ||
** pGP563 (N-terminal His-tag, in [[pWH844]]), available in [[Stülke]] lab | ** pGP563 (N-terminal His-tag, in [[pWH844]]), available in [[Stülke]] lab | ||
** pGP93 (N-terminal Strep-tag, purification from ''B. subtilis'', for [[SPINE]], in [[pGP380]]), available in [[Stülke]] lab | ** pGP93 (N-terminal Strep-tag, purification from ''B. subtilis'', for [[SPINE]], in [[pGP380]]), available in [[Stülke]] lab | ||
| − | pGP1500 (expression in ''B. subtilis'', in [[pBQ200]]), available in [[Stülke]] lab | + | ** pGP1500 (expression in ''B. subtilis'', in [[pBQ200]]), available in [[Stülke]] lab |
* '''lacZ fusion:''' | * '''lacZ fusion:''' | ||
Revision as of 10:42, 7 July 2009
- Description: enolase, glycolytic/ gluconeogenic enzyme
| Gene name | eno |
| Synonyms | |
| Essential | yes |
| Product | enolase |
| Function | enzyme in glycolysis/ gluconeogenesis |
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism | |
| MW, pI | 46,4 kDa, 4.49 |
| Gene length, protein length | 1290 bp, 430 amino acids |
| Immediate neighbours | pgm, yvgK |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 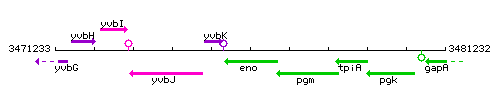
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU33900
Phenotypes of a mutant
- Essential PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 2-phospho-D-glycerate = phosphoenolpyruvate + H2O (according to Swiss-Prot) 2-phospho-D-glycerate = phosphoenolpyruvate + H(2)O
- Protein family: enolase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information: Reversible Michaelis-Menten PubMed
- Domains:
- substrate binding domain (366–369)
- Cofactor(s): Mg2+
- Effectors of protein activity:
- Inhibited by EDTA PubMed
Database entries
- Structure: 3ES8 (from Oceanobacillus iheyensis, complex with Mg(2+) and malate)
- Swiss prot entry: P37869
- KEGG entry: [3]
- E.C. number: 4.2.1.11
Additional information
There are indications that this enzyme is an octamer PubMed
Expression and regulation
- Sigma factor: SigA
- Regulation: expression activated by glucose (3.3 fold) PubMed
- Additional information:
Biological materials
- Mutant: GP698 (cat), available in Stülke lab
- Expression vector:
- lacZ fusion:
- GFP fusion: pHT315-yfp-eno, available in Mijakovic lab
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody: available in Stülke lab
Labs working on this gene/protein
Jörg Stülke, University of Göttingen, Germany Homepage
Your additional remarks
References