Difference between revisions of "TrpE"
| Line 35: | Line 35: | ||
=== Basic information === | === Basic information === | ||
| − | * '''Locus tag:''' | + | * '''Locus tag:''' BSU22680 |
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| Line 80: | Line 80: | ||
* '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/P03963 P03963] | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/P03963 P03963] | ||
| − | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+ | + | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+BSU22680] |
* '''E.C. number:''' [http://www.expasy.org/enzyme/4.1.3.27 4.1.3.27] | * '''E.C. number:''' [http://www.expasy.org/enzyme/4.1.3.27 4.1.3.27] | ||
Revision as of 10:11, 3 June 2009
- Description: anthranilate synthase
| Gene name | trpE |
| Synonyms | |
| Essential | no |
| Product | anthranilate synthase |
| Function | biosynthesis of tryptophan |
| MW, pI | 57 kDa, 5.246 |
| Gene length, protein length | 1545 bp, 515 aa |
| Immediate neighbours | trpD, aroH |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 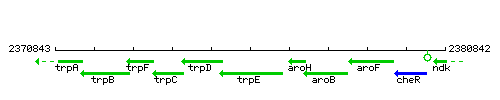
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU22680
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Chorismate + L-glutamine = anthranilate + pyruvate + L-glutamate (according to Swiss-Prot)
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity: subject to feedback inhibition by tryptophan PubMed
- Interactions:
- Localization:
Database entries
- Structure:
- Swiss prot entry: P03963
- KEGG entry: [3]
- E.C. number: 4.1.3.27
Additional information
Expression and regulation
- Regulatory mechanism: TRAP: binding to the mRNA in the presence of tryptophan, this results in transcription termination PubMed
- Additional information: subject to feedback inhibition by tryptophan PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Gintaras Deikus, Paul Babitzke, David H Bechhofer
Recycling of a regulatory protein by degradation of the RNA to which it binds.
Proc Natl Acad Sci U S A: 2004, 101(9);2747-51
[PubMed:14976255]
[WorldCat.org]
[DOI]
(P p)
J Otridge, P Gollnick
MtrB from Bacillus subtilis binds specifically to trp leader RNA in a tryptophan-dependent manner.
Proc Natl Acad Sci U S A: 1993, 90(1);128-32
[PubMed:8419914]
[WorldCat.org]
[DOI]
(P p)
P Babitzke, P Gollnick, C Yanofsky
The mtrAB operon of Bacillus subtilis encodes GTP cyclohydrolase I (MtrA), an enzyme involved in folic acid biosynthesis, and MtrB, a regulator of tryptophan biosynthesis.
J Bacteriol: 1992, 174(7);2059-64
[PubMed:1551827]
[WorldCat.org]
[DOI]
(P p)
H Shimotsu, M I Kuroda, C Yanofsky, D J Henner
Novel form of transcription attenuation regulates expression the Bacillus subtilis tryptophan operon.
J Bacteriol: 1986, 166(2);461-71
[PubMed:2422155]
[WorldCat.org]
[DOI]
(P p)
D J Henner, L Band, H Shimotsu
Nucleotide sequence of the Bacillus subtilis tryptophan operon.
Gene: 1985, 34(2-3);169-77
[PubMed:3924737]
[WorldCat.org]
[DOI]
(P p)
H Shimotsu, D J Henner
Characterization of the Bacillus subtilis tryptophan promoter region.
Proc Natl Acad Sci U S A: 1984, 81(20);6315-9
[PubMed:6436812]
[WorldCat.org]
[DOI]
(P p)
- Author1, Author2 & Author3 (year) Title Journal volume: page-page. PubMed