Difference between revisions of "RocF"
| Line 123: | Line 123: | ||
* '''Additional information:''' | * '''Additional information:''' | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium): 126 {{PubMed|24696501}} | ||
| + | ** number of protein molecules per cell (complex medium with amino acids, without glucose): 13789 {{PubMed|24696501}} | ||
=Biological materials = | =Biological materials = | ||
Revision as of 09:57, 17 April 2014
- Description: arginase
| Gene name | rocF |
| Synonyms | |
| Essential | no |
| Product | arginase |
| Function | arginine utilization |
| Gene expression levels in SubtiExpress: rocF | |
| Metabolic function and regulation of this protein in SubtiPathways: rocF | |
| MW, pI | 32 kDa, 4.932 |
| Gene length, protein length | 888 bp, 296 aa |
| Immediate neighbours | phrG, rocE |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 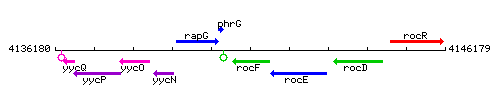
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
AhrC regulon, CodY regulon, RocR regulon, SigL regulon, Spo0A regulon
The gene
Basic information
- Locus tag: BSU40320
Phenotypes of a mutant
Database entries
- BsubCyc: BSU40320
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: L-arginine + H2O = L-ornithine + urea (according to Swiss-Prot)
- Protein family: arginase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification: phosphorylated on Ser-68 PubMed
- Effectors of protein activity:
Database entries
- BsubCyc: BSU40320
- UniProt: P39138
- KEGG entry: [3]
- E.C. number: 3.5.3.1
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant: GP655, aphA3, available in the Stülke lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Jin-Ju Yu, Ki-Bum Park, Su-Gon Kim, Suk-Heung Oh
Expression, purification, and biochemical properties of arginase from Bacillus subtilis 168.
J Microbiol: 2013, 51(2);222-8
[PubMed:23625224]
[WorldCat.org]
[DOI]
(I p)
Boumediene Soufi, Chanchal Kumar, Florian Gnad, Matthias Mann, Ivan Mijakovic, Boris Macek
Stable isotope labeling by amino acids in cell culture (SILAC) applied to quantitative proteomics of Bacillus subtilis.
J Proteome Res: 2010, 9(7);3638-46
[PubMed:20509597]
[WorldCat.org]
[DOI]
(I p)
Virginie Molle, Masaya Fujita, Shane T Jensen, Patrick Eichenberger, José E González-Pastor, Jun S Liu, Richard Losick
The Spo0A regulon of Bacillus subtilis.
Mol Microbiol: 2003, 50(5);1683-701
[PubMed:14651647]
[WorldCat.org]
[DOI]
(P p)
Virginie Molle, Yoshiko Nakaura, Robert P Shivers, Hirotake Yamaguchi, Richard Losick, Yasutaro Fujita, Abraham L Sonenshein
Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis.
J Bacteriol: 2003, 185(6);1911-22
[PubMed:12618455]
[WorldCat.org]
[DOI]
(P p)
M C Bewley, P D Jeffrey, M L Patchett, Z F Kanyo, E N Baker
Crystal structures of Bacillus caldovelox arginase in complex with substrate and inhibitors reveal new insights into activation, inhibition and catalysis in the arginase superfamily.
Structure: 1999, 7(4);435-48
[PubMed:10196128]
[WorldCat.org]
[DOI]
(P p)
R Gardan, G Rapoport, M Débarbouillé
Expression of the rocDEF operon involved in arginine catabolism in Bacillus subtilis.
J Mol Biol: 1995, 249(5);843-56
[PubMed:7540694]
[WorldCat.org]
[DOI]
(P p)