Difference between revisions of "SpoIVFB"
(→Reviews) |
|||
| Line 97: | Line 97: | ||
* '''[[Localization]]:''' | * '''[[Localization]]:''' | ||
** integral membrane protein {{PubMed|11959848}} | ** integral membrane protein {{PubMed|11959848}} | ||
| − | ** | + | ** mother cell membrane {{PubMed|24243021}} |
=== Database entries === | === Database entries === | ||
| Line 150: | Line 150: | ||
==Original Publications== | ==Original Publications== | ||
| − | <pubmed>11959848,9501233,12940997,1577688,12060714,9078383,1942049,10611287,15383836 16818230 19805276 15699190 23585539 23995631 15087499</pubmed> | + | <pubmed>11959848,9501233,12940997,1577688,12060714,9078383,1942049,10611287,15383836 16818230 19805276 15699190 23585539 23995631 15087499 24243021 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 10:02, 3 December 2013
- Description: intramembrane metalloprotease, processing of pro-sigma-K to active SigK
| Gene name | spoIVFB |
| Synonyms | |
| Essential | no |
| Product | intramembrane metalloprotease |
| Function | processing of pro-sigma-K to active SigK |
| Gene expression levels in SubtiExpress: spoIVFB | |
| Interactions involving this protein in SubtInteract: SpoIVFB | |
| MW, pI | 33 kDa, 8.483 |
| Gene length, protein length | 864 bp, 288 aa |
| Immediate neighbours | rplU, spoIVFA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 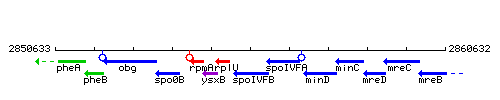
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed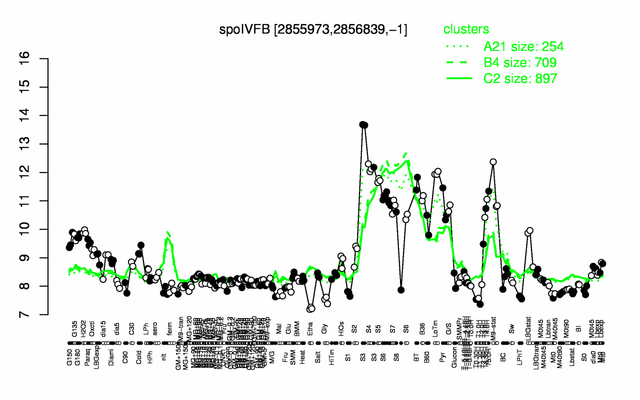
| |
Contents
Categories containing this gene/protein
sigma factors and their control, proteolysis, sporulation proteins, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU27970
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Protein family: peptidase M50B family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- C-terminal cystathionine-beta-synthase (CBS) domain, this domain binds ATP PubMed
- Modification:
- Cofactor(s):
- Effectors of protein activity: ATP regulates substrate access to the active site and renders cleavage sensitive to the cellular energy level PubMed
Database entries
- Structure:
- UniProt: P26937
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Lee Kroos, Yoshinori Akiyama
Biochemical and structural insights into intramembrane metalloprotease mechanisms.
Biochim Biophys Acta: 2013, 1828(12);2873-85
[PubMed:24099006]
[WorldCat.org]
[DOI]
(P p)
Noël Molière, Kürşad Turgay
General and regulatory proteolysis in Bacillus subtilis.
Subcell Biochem: 2013, 66;73-103
[PubMed:23479438]
[WorldCat.org]
[DOI]
(P p)
Gu Chen, Xu Zhang
New insights into S2P signaling cascades: regulation, variation, and conservation.
Protein Sci: 2010, 19(11);2015-30
[PubMed:20836086]
[WorldCat.org]
[DOI]
(I p)
Michael S Wolfe
Intramembrane-cleaving proteases.
J Biol Chem: 2009, 284(21);13969-73
[PubMed:19189971]
[WorldCat.org]
[DOI]
(P p)
Original Publications