Difference between revisions of "ExoA"
(→References) |
|||
| Line 35: | Line 35: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | |||
| − | |||
| − | |||
| − | |||
<br/><br/> | <br/><br/> | ||
| Line 121: | Line 117: | ||
* '''Mutant:''' | * '''Mutant:''' | ||
| − | + | ** GP898 (''exoA''::''kan''), available in [[Jörg Stülke]]'s lab | |
| − | ** GP898 (''exoA''::''kan''), available in [[Stülke]] lab | + | ** GP1503 (''exoA''::''kan'', ''nfo''::''cat''), available in [[Jörg Stülke]]'s lab |
| − | ** GP1503 (''exoA''::''kan'', ''nfo''::''cat''), available in [[Stülke]] lab | ||
* '''Expression vector:''' | * '''Expression vector:''' | ||
| Line 143: | Line 138: | ||
<pubmed> 22933559 </pubmed> | <pubmed> 22933559 </pubmed> | ||
== Original publications == | == Original publications == | ||
| − | <pubmed>18203828,16237020, 19930460, 10540738 21441501</pubmed> | + | <pubmed>18203828,16237020, 19930460, 10540738 21441501 24123749 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 14:25, 21 October 2013
- Description: apurinic/apyrimidinic endonuclease, multifunctional DNA-repair enzyme, important for spore dormance
| Gene name | exoA |
| Synonyms | |
| Essential | no |
| Product | apurinic/apyrimidinic endonuclease |
| Function | repair of oxidative DNA damage in spores |
| Gene expression levels in SubtiExpress: exoA | |
| MW, pI | 29 kDa, 4.592 |
| Gene length, protein length | 756 bp, 252 aa |
| Immediate neighbours | ccpB, rpsR |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 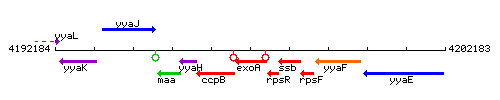
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
DNA repair/ recombination, sporulation proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU40880
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Exonucleolytic cleavage in the 3'- to 5'-direction to yield nucleoside 5'-phosphates (according to Swiss-Prot)
- Protein family: DNA repair enzymes AP/exoA family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- cytoplasm (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: P37454
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- GP898 (exoA::kan), available in Jörg Stülke's lab
- GP1503 (exoA::kan, nfo::cat), available in Jörg Stülke's lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Justin S Lenhart, Jeremy W Schroeder, Brian W Walsh, Lyle A Simmons
DNA repair and genome maintenance in Bacillus subtilis.
Microbiol Mol Biol Rev: 2012, 76(3);530-64
[PubMed:22933559]
[WorldCat.org]
[DOI]
(I p)
Original publications