Difference between revisions of "QueF"
(→Extended information on the protein) |
|||
| Line 42: | Line 42: | ||
* '''Locus tag:''' BSU13750 | * '''Locus tag:''' BSU13750 | ||
| + | |||
| + | [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=queF_1441291_1441788_1 Expression] | ||
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
Revision as of 08:35, 25 January 2012
- Description: nitrile reductase, synthesis of the modified ribonucleotide queuosine
| Gene name | queF |
| Synonyms | ykvM |
| Essential | no |
| Product | nitrile reductase |
| Function | tRNA modification |
| MW, pI | 19 kDa, 4.927 |
| Gene length, protein length | 495 bp, 165 aa |
| Immediate neighbours | queE, ykvN |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 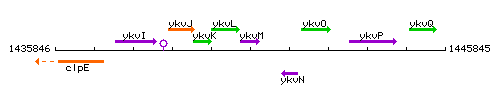
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU13750
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 7-aminomethyl-7-carbaguanine + 2 NADP+ = 7-cyano-7-carbaguanine + 2 NADPH (according to Swiss-Prot)
- Protein family: QueF type 1 subfamily (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: O31678
- KEGG entry: [2]
- E.C. number: 1.7.1.13
Additional information
Expression and regulation
- Regulation:
- repressed in the presence of queuosine (preQ1 riboswitch) PubMed
- Regulatory mechanism:
- preQ1 riboswitch: transcriptional antitermination in the absence of queuosine PubMed
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Mijeong Kang, Robert Peterson, Juli Feigon
Structural Insights into riboswitch control of the biosynthesis of queuosine, a modified nucleotide found in the anticodon of tRNA.
Mol Cell: 2009, 33(6);784-90
[PubMed:19285444]
[WorldCat.org]
[DOI]
(I p)
Bobby W K Lee, Steven G Van Lanen, Dirk Iwata-Reuyl
Mechanistic studies of Bacillus subtilis QueF, the nitrile oxidoreductase involved in queuosine biosynthesis.
Biochemistry: 2007, 46(44);12844-54
[PubMed:17929836]
[WorldCat.org]
[DOI]
(P p)
Adam Roth, Wade C Winkler, Elizabeth E Regulski, Bobby W K Lee, Jinsoo Lim, Inbal Jona, Jeffrey E Barrick, Ankita Ritwik, Jane N Kim, Rüdiger Welz, Dirk Iwata-Reuyl, Ronald R Breaker
A riboswitch selective for the queuosine precursor preQ1 contains an unusually small aptamer domain.
Nat Struct Mol Biol: 2007, 14(4);308-17
[PubMed:17384645]
[WorldCat.org]
[DOI]
(P p)
Manal A Swairjo, Robert R Reddy, Bobby Lee, Steven G Van Lanen, Shannon Brown, Valérie de Crécy-Lagard, Dirk Iwata-Reuyl, Paul Schimmel
Crystallization and preliminary X-ray characterization of the nitrile reductase QueF: a queuosine-biosynthesis enzyme.
Acta Crystallogr Sect F Struct Biol Cryst Commun: 2005, 61(Pt 10);945-8
[PubMed:16511203]
[WorldCat.org]
[DOI]
(I p)
Steven G Van Lanen, John S Reader, Manal A Swairjo, Valérie de Crécy-Lagard, Bobby Lee, Dirk Iwata-Reuyl
From cyclohydrolase to oxidoreductase: discovery of nitrile reductase activity in a common fold.
Proc Natl Acad Sci U S A: 2005, 102(12);4264-9
[PubMed:15767583]
[WorldCat.org]
[DOI]
(P p)
John S Reader, David Metzgar, Paul Schimmel, Valérie de Crécy-Lagard
Identification of four genes necessary for biosynthesis of the modified nucleoside queuosine.
J Biol Chem: 2004, 279(8);6280-5
[PubMed:14660578]
[WorldCat.org]
[DOI]
(P p)