Difference between revisions of "McsB"
(→Expression and regulation) |
|||
| Line 1: | Line 1: | ||
| − | * '''Description:''' protein arginine kinase | + | * '''Description:''' protein arginine kinase, modulator of [[CtsR]]-dependent repression <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 54: | Line 54: | ||
=== Basic information/ Evolution === | === Basic information/ Evolution === | ||
| − | * '''Catalyzed reaction/ biological activity:''' | + | * '''Catalyzed reaction/ biological activity:''' targets non-functional CtsR for degradation by [[ClpP]]/[[ClpC]] {{PubMed|20852588}} |
| + | |||
* '''Protein family:''' ATP:guanido phosphotransferase family (according to Swiss-Prot) | * '''Protein family:''' ATP:guanido phosphotransferase family (according to Swiss-Prot) | ||
| Line 66: | Line 67: | ||
* '''Domains:''' | * '''Domains:''' | ||
| − | * '''Modification:''' | + | * '''Modification:''' autophosphorylation {{PubMed|20852588}}, dephosphorylation by [[YwlE]] {{PubMed|20852588}} |
* '''Cofactor(s):''' | * '''Cofactor(s):''' | ||
| Line 72: | Line 73: | ||
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| − | * '''Interactions:''' [[McsA]]-[[McsB]]-[[CtsR]], [[McsB]]-[[CtsR]], | + | * '''Interactions:''' |
| + | ** [[McsA]]-[[McsB]]-[[CtsR]], [[McsB]]-[[CtsR]] {{PubMed|20852588}}, [[McsA]]-[[McsB]] | ||
| + | ** [[McsB]]-[[YwlE]] {{PubMed|20852588}} | ||
| + | ** [[McsB]]-[[ClpP]]/[[ClpC]] (rapid degradation of non-phosphorylated McsB) {{PubMed|20852588}} | ||
* '''Localization:''' | * '''Localization:''' | ||
| Line 127: | Line 131: | ||
<pubmed> 19609260 </pubmed> | <pubmed> 19609260 </pubmed> | ||
==Original Publications== | ==Original Publications== | ||
| − | <pubmed>17380125,16163393,,17380125,11179229,9987115,8793870,16163393,16497325,19226326,,9987115, 19498169 ,11544224, </pubmed> | + | <pubmed>17380125,16163393,,17380125,11179229,9987115,8793870,16163393,16497325,19226326,,9987115, 19498169 ,11544224, 20852588 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 08:56, 26 October 2010
- Description: protein arginine kinase, modulator of CtsR-dependent repression
| Gene name | mcsB |
| Synonyms | yacI |
| Essential | no |
| Product | protein arginine kinase |
| Function | control of CtsR activity |
| Metabolic function and regulation of this protein in SubtiPathways: Stress | |
| MW, pI | 40 kDa, 5.068 |
| Gene length, protein length | 1089 bp, 363 aa |
| Immediate neighbours | mcsA, clpC |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 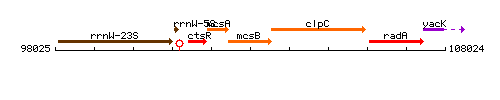
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU00850
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: targets non-functional CtsR for degradation by ClpP/ClpC PubMed
- Protein family: ATP:guanido phosphotransferase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure:
- UniProt: P37570
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original Publications
Alexander K W Elsholz, Stephan Michalik, Daniela Zühlke, Michael Hecker, Ulf Gerth
CtsR, the Gram-positive master regulator of protein quality control, feels the heat.
EMBO J: 2010, 29(21);3621-9
[PubMed:20852588]
[WorldCat.org]
[DOI]
(I p)
Jakob Fuhrmann, Andreas Schmidt, Silvia Spiess, Anita Lehner, Kürsad Turgay, Karl Mechtler, Emmanuelle Charpentier, Tim Clausen
McsB is a protein arginine kinase that phosphorylates and inhibits the heat-shock regulator CtsR.
Science: 2009, 324(5932);1323-7
[PubMed:19498169]
[WorldCat.org]
[DOI]
(I p)
Jeanette Hahn, Naomi Kramer, Kenneth Briley, David Dubnau
McsA and B mediate the delocalization of competence proteins from the cell poles of Bacillus subtilis.
Mol Microbiol: 2009, 72(1);202-15
[PubMed:19226326]
[WorldCat.org]
[DOI]
(I p)
Janine Kirstein, David A Dougan, Ulf Gerth, Michael Hecker, Kürşad Turgay
The tyrosine kinase McsB is a regulated adaptor protein for ClpCP.
EMBO J: 2007, 26(8);2061-70
[PubMed:17380125]
[WorldCat.org]
[DOI]
(P p)
Stephanie T Wang, Barbara Setlow, Erin M Conlon, Jessica L Lyon, Daisuke Imamura, Tsutomu Sato, Peter Setlow, Richard Losick, Patrick Eichenberger
The forespore line of gene expression in Bacillus subtilis.
J Mol Biol: 2006, 358(1);16-37
[PubMed:16497325]
[WorldCat.org]
[DOI]
(P p)
Janine Kirstein, Daniela Zühlke, Ulf Gerth, Kürşad Turgay, Michael Hecker
A tyrosine kinase and its activator control the activity of the CtsR heat shock repressor in B. subtilis.
EMBO J: 2005, 24(19);3435-45
[PubMed:16163393]
[WorldCat.org]
[DOI]
(P p)
A Petersohn, M Brigulla, S Haas, J D Hoheisel, U Völker, M Hecker
Global analysis of the general stress response of Bacillus subtilis.
J Bacteriol: 2001, 183(19);5617-31
[PubMed:11544224]
[WorldCat.org]
[DOI]
(P p)
E Krüger, D Zühlke, E Witt, H Ludwig, M Hecker
Clp-mediated proteolysis in Gram-positive bacteria is autoregulated by the stability of a repressor.
EMBO J: 2001, 20(4);852-63
[PubMed:11179229]
[WorldCat.org]
[DOI]
(P p)
I Derré, G Rapoport, T Msadek
CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in gram-positive bacteria.
Mol Microbiol: 1999, 31(1);117-31
[PubMed:9987115]
[WorldCat.org]
[DOI]
(P p)
E Krüger, T Msadek, M Hecker
Alternate promoters direct stress-induced transcription of the Bacillus subtilis clpC operon.
Mol Microbiol: 1996, 20(4);713-23
[PubMed:8793870]
[WorldCat.org]
[DOI]
(P p)