Difference between revisions of "AddA"
(→Original publications) |
(→Original publications) |
||
| Line 148: | Line 148: | ||
==Original publications== | ==Original publications== | ||
| − | <pubmed>,8387145,15610857,7746142, 19129187 15009890 10756102 9781875 16632468 17570399 20350930 21809208 21821766 22307084 24682829 24670664 22383849 21071401 8752329 </pubmed> | + | <pubmed>,8387145,15610857,7746142, 19129187 15009890 10756102 9781875 16632468 17570399 20350930 21809208 21821766 22307084 24682829 24670664 22383849 21071401 8752329 8752323 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 13:03, 26 June 2014
- Description: ATP-dependent deoxyribonuclease (subunit A)
| Gene name | addA |
| Synonyms | recE5 |
| Essential | no |
| Product | ATP-dependent deoxyribonuclease (subunit A)) |
| Function | DNA repair/ recombination |
| Gene expression levels in SubtiExpress: addA | |
| Interactions involving this protein in SubtInteract: AddA | |
| MW, pI | 140 kDa, 5.127 |
| Gene length, protein length | 3696 bp, 1232 aa |
| Immediate neighbours | addB, sbcD |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 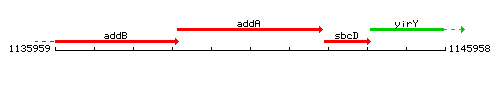
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed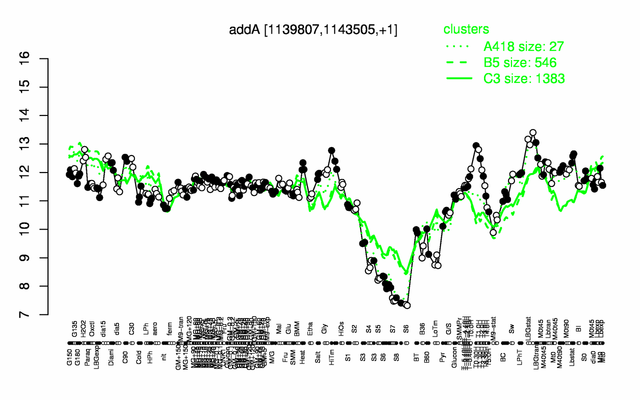
| |
Contents
Categories containing this gene/protein
DNA repair/ recombination, genetic competence
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU10630
Phenotypes of a mutant
Database entries
- BsubCyc: BSU10630
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
- A mutation was found in this gene after evolution under relaxed selection for sporulation PubMed
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- the enzyme is functional as a heterodimer of the AddA and AddB subunits, that it is a rapid and processive DNA helicase, and that it catalyses DNA unwinding using one single-stranded DNA motor of 3'→5' polarity located in the AddA subunit PubMed
- the AddB subunit contains a second putative ATP-binding pocket, but this does not contribute to the observed helicase activity and may instead be involved in the recognition of recombination hotspot sequences PubMed
- Protein family: uvrD-like helicase C-terminal domain (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- BsubCyc: BSU10630
- UniProt: P23478
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
- number of protein molecules per cell (minimal medium with glucose and ammonium): 147 PubMed
Biological materials
- Mutant: GP1106 (addAB, spc), available in Stülke lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
- Mark Dillingham, Bristol, U.K. (homepage)
Your additional remarks
References
Reviews
Dale B Wigley
Bacterial DNA repair: recent insights into the mechanism of RecBCD, AddAB and AdnAB.
Nat Rev Microbiol: 2013, 11(1);9-13
[PubMed:23202527]
[WorldCat.org]
[DOI]
(I p)
Justin S Lenhart, Jeremy W Schroeder, Brian W Walsh, Lyle A Simmons
DNA repair and genome maintenance in Bacillus subtilis.
Microbiol Mol Biol Rev: 2012, 76(3);530-64
[PubMed:22933559]
[WorldCat.org]
[DOI]
(I p)
Joseph T P Yeeles, Mark S Dillingham
The processing of double-stranded DNA breaks for recombinational repair by helicase-nuclease complexes.
DNA Repair (Amst): 2010, 9(3);276-85
[PubMed:20116346]
[WorldCat.org]
[DOI]
(I p)
Original publications