Difference between revisions of "YqeV"
| Line 54: | Line 54: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU25430&redirect=T BSU25430] | ||
* '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/lepA-hemN-hrcA-grpE-dnaKJ-yqeTUV.html] | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/lepA-hemN-hrcA-grpE-dnaKJ-yqeTUV.html] | ||
| Line 91: | Line 92: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU25430&redirect=T BSU25430] | ||
* '''Structure:''' | * '''Structure:''' | ||
Revision as of 14:15, 2 April 2014
- Description: tRNA methylthiotransferase
| Gene name | yqeV |
| Synonyms | mtaB |
| Essential | no |
| Product | tRNA methylthiotransferase |
| Function | tRNA maturation |
| Gene expression levels in SubtiExpress: yqeV | |
| MW, pI | 51 kDa, 5.242 |
| Gene length, protein length | 1353 bp, 451 aa |
| Immediate neighbours | yqeW, yqeU |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 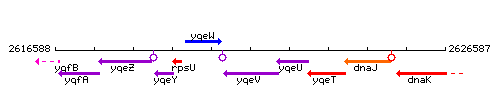
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
translation, heat shock proteins, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU25430
Phenotypes of a mutant
Database entries
- BsubCyc: BSU25430
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: modification of N(6)-threonylcarbamoyladenosine (t(6)A) to 2-methylthio-N(6)-threonylcarbamoyladenosine (ms(2)t(6)A) in tRNA PubMed
- Protein family: YqeV subfamily (according to Swiss-Prot)
- Paralogous protein(s): YmcB
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s): 2 (4Fe-4S) clusters PubMed
- Effectors of protein activity:
- Localization: cell membrane (according to Swiss-Prot)
Database entries
- BsubCyc: BSU25430
- Structure:
- UniProt: P54462
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Simon Arragain, Samuel K Handelman, Farhad Forouhar, Fan-Yan Wei, Kazuhito Tomizawa, John F Hunt, Thierry Douki, Marc Fontecave, Etienne Mulliez, Mohamed Atta
Identification of eukaryotic and prokaryotic methylthiotransferase for biosynthesis of 2-methylthio-N6-threonylcarbamoyladenosine in tRNA.
J Biol Chem: 2010, 285(37);28425-33
[PubMed:20584901]
[WorldCat.org]
[DOI]
(I p)
Brian P Anton, Susan P Russell, Jason Vertrees, Simon Kasif, Elisabeth A Raleigh, Patrick A Limbach, Richard J Roberts
Functional characterization of the YmcB and YqeV tRNA methylthiotransferases of Bacillus subtilis.
Nucleic Acids Res: 2010, 38(18);6195-205
[PubMed:20472640]
[WorldCat.org]
[DOI]
(I p)
G Homuth, A Mogk, W Schumann
Post-transcriptional regulation of the Bacillus subtilis dnaK operon.
Mol Microbiol: 1999, 32(6);1183-97
[PubMed:10383760]
[WorldCat.org]
[DOI]
(P p)
G Homuth, S Masuda, A Mogk, Y Kobayashi, W Schumann
The dnaK operon of Bacillus subtilis is heptacistronic.
J Bacteriol: 1997, 179(4);1153-64
[PubMed:9023197]
[WorldCat.org]
[DOI]
(P p)
G Yuan, S L Wong
Isolation and characterization of Bacillus subtilis groE regulatory mutants: evidence for orf39 in the dnaK operon as a repressor gene in regulating the expression of both groE and dnaK.
J Bacteriol: 1995, 177(22);6462-8
[PubMed:7592421]
[WorldCat.org]
[DOI]
(P p)