Difference between revisions of "YvcL"
(→Phenotypes of a mutant) |
|||
| Line 51: | Line 51: | ||
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| − | * a ''[[yvcL]] [[zapA]]'' double mutant grows poorly, and the cells are filamentous {{PubMed|24097947}} | + | * a ''[[yvcL]] [[zapA]]'' double mutant grows poorly, and the cells are filamentous (due to a defect in Z ring assembly) {{PubMed|24097947}} |
| + | * a ''[[yvcL]] [[ezrA]]'' double mutant grows poorly, and the cells are filamentous {{PubMed|24097947}} | ||
| + | * a ''[[yvcL]] [[minC]]-[[minD]]'' mutant grows poorly, and the cells are filamentous {{PubMed|24097947}} | ||
| + | * a ''[[yvcL]] [[noc]]'' double mutant grows poorly, and the cells are filamentous {{PubMed|24097947}} | ||
* the defects of the ''[[yvcL]] [[zapA]]'' double mutant can be suppressed by inactivation of either ''[[gtaB]]'', ''[[pgcA]]'', or ''[[ugtP]]'' {{PubMed|24097947}} | * the defects of the ''[[yvcL]] [[zapA]]'' double mutant can be suppressed by inactivation of either ''[[gtaB]]'', ''[[pgcA]]'', or ''[[ugtP]]'' {{PubMed|24097947}} | ||
Revision as of 18:03, 21 December 2013
- Description: WhiA family protein, involved in Z ring assembly, putative transcription factor
| Gene name | yvcL |
| Synonyms | |
| Essential | no |
| Product | cell division protein |
| Function | control of Z ring asembly |
| Gene expression levels in SubtiExpress: yvcL | |
| MW, pI | 36 kDa, 9.479 |
| Gene length, protein length | 948 bp, 316 aa |
| Immediate neighbours | crh, yvcK |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 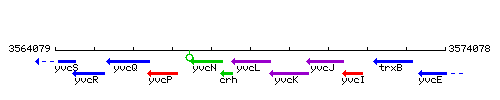
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
cell division, transcription factors and their control
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU34750
Phenotypes of a mutant
- a yvcL zapA double mutant grows poorly, and the cells are filamentous (due to a defect in Z ring assembly) PubMed
- a yvcL ezrA double mutant grows poorly, and the cells are filamentous PubMed
- a yvcL minC-minD mutant grows poorly, and the cells are filamentous PubMed
- a yvcL noc double mutant grows poorly, and the cells are filamentous PubMed
- the defects of the yvcL zapA double mutant can be suppressed by inactivation of either gtaB, pgcA, or ugtP PubMed
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: WhiA family PubMed
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
- Localization:
- localizes to the nucleoid PubMed
Database entries
- Structure:
- UniProt: O06975
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant: GP231 (amyE::PrpsJ-lacZ, yvcL::cat) available in Jörg Stülke's lab; plasmid pGP1468 was used to disrupt the gene.
- Expression vector: pGP1469 for expression in B. subtilis (in pGP382, with C-terminal STREP-TAG), available in Jörg Stülke's lab
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Jörg Stülke's and Boris Görke's labs
- Antibody: available in Leendert Hamoen's lab PubMed
Labs working on this gene/protein
Your additional remarks
WhiA orthologues are present in all Gram-positive bacteria and best characterized by the WhiA sporulation control factor in Streptomyces coelicolor. They are thought to act as genetic regulators that have evolved from LAGLIDADG homing endonucleases (LHE).
References