Difference between revisions of "RbsR"
| Line 50: | Line 50: | ||
| + | |||
| + | = Categories containing this gene/protein = | ||
| + | {{SubtiWiki category|[[utilization of specific carbon sources]]}}, | ||
| + | {{SubtiWiki category|[[transcription factors and their control]]}} | ||
=The protein= | =The protein= | ||
Revision as of 19:15, 30 November 2010
- Description: transcriptional repressor of the ribose operon
| Gene name | rbsR |
| Synonyms | |
| Essential | no |
| Product | transcriptional repressor (LacI family) |
| Function | regulation of ribose utilization |
| Metabolic function and regulation of this protein in SubtiPathways: Sugar catabolism | |
| MW, pI | 35 kDa, 5.577 |
| Gene length, protein length | 978 bp, 326 aa |
| Immediate neighbours | capB, rbsK |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 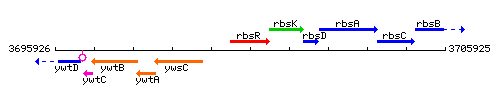
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU35910
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
Categories containing this gene/protein
utilization of specific carbon sources, transcription factors and their control
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: LacI family
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
Database entries
- Structure:
- UniProt: P36944
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Le Thi Tam, Christine Eymann, Dirk Albrecht, Rabea Sietmann, Frieder Schauer, Michael Hecker, Haike Antelmann
Differential gene expression in response to phenol and catechol reveals different metabolic activities for the degradation of aromatic compounds in Bacillus subtilis.
Environ Microbiol: 2006, 8(8);1408-27
[PubMed:16872404]
[WorldCat.org]
[DOI]
(P p)
Wolfgang Müller, Nicola Horstmann, Wolfgang Hillen, Heinrich Sticht
The transcription regulator RbsR represents a novel interaction partner of the phosphoprotein HPr-Ser46-P in Bacillus subtilis.
FEBS J: 2006, 273(6);1251-61
[PubMed:16519689]
[WorldCat.org]
[DOI]
(P p)
Hans-Matti Blencke, Georg Homuth, Holger Ludwig, Ulrike Mäder, Michael Hecker, Jörg Stülke
Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways.
Metab Eng: 2003, 5(2);133-49
[PubMed:12850135]
[WorldCat.org]
[DOI]
(P p)
M A Strauch
AbrB modulates expression and catabolite repression of a Bacillus subtilis ribose transport operon.
J Bacteriol: 1995, 177(23);6727-31
[PubMed:7592460]
[WorldCat.org]
[DOI]
(P p)
K Woodson, K M Devine
Analysis of a ribose transport operon from Bacillus subtilis.
Microbiology (Reading): 1994, 140 ( Pt 8);1829-38
[PubMed:7921236]
[WorldCat.org]
[DOI]
(P p)
M O'Reilly, K Woodson, B C Dowds, K M Devine
The citrulline biosynthetic operon, argC-F, and a ribose transport operon, rbs, from Bacillus subtilis are negatively regulated by Spo0A.
Mol Microbiol: 1994, 11(1);87-98
[PubMed:7511775]
[WorldCat.org]
[DOI]
(P p)